Data Fundamentals
Load, clean, reshape, and merge your data
Think of Data as Excel Spreadsheets
The datasets we work with in Stata and R are just tables—exactly like Excel spreadsheets. Each row is an observation (a person, a country-year, a transaction), and each column is a variable (age, GDP, price). If you've used Excel, you already understand the basic structure. The difference is that instead of clicking and dragging, we write code to manipulate these tables—which makes our work reproducible and scalable to millions of rows.
Reading & Writing Files
Loading Data
* Load Stata dataset
use "data.dta", clear
* Load CSV file
import delimited "data.csv", clear
* Load Excel file
import excel "data.xlsx", sheet("Sheet1") firstrow clearlibrary(haven) # For Stata files
library(readr) # For CSV files
library(readxl) # For Excel files
# Load Stata dataset
data <- read_dta("data.dta")
# Load CSV file
data <- read_csv("data.csv")
# Load Excel file
data <- read_excel("data.xlsx", sheet = "Sheet1")import pandas as pd
# Load Stata dataset
data = pd.read_stata("data.dta")
# Load CSV file
data = pd.read_csv("data.csv")
# Load Excel file
data = pd.read_excel("data.xlsx", sheet_name="Sheet1")Saving Data
* Save as Stata dataset
save "cleaned_data.dta", replace
* Export to CSV
export delimited "cleaned_data.csv", replace# Save as Stata dataset
write_dta(data, "cleaned_data.dta")
# Export to CSV
write_csv(data, "cleaned_data.csv")# Save as Stata dataset
data.to_stata("cleaned_data.dta", write_index=False)
# Export to CSV
data.to_csv("cleaned_data.csv", index=False)Save Intermediate Files
Save your data after each major step with numbered prefixes. When something breaks at step 5, you can start from step 4 instead of re-running everything:
01_imported.dta02_cleaned.dta03_merged.dta
Data Cleaning
Inspecting Your Data
* Overview of dataset
describe
* Summary statistics
summarize
* First few rows
list in 1/10
* Check for missing values
misstable summarize# Overview of dataset
str(data)
glimpse(data)
# Summary statistics
summary(data)
# First few rows
head(data, 10)
# Check for missing values
colSums(is.na(data))# Overview of dataset
data.info()
data.dtypes
# Summary statistics
data.describe()
# First few rows
data.head(10)
# Check for missing values
data.isna().sum()Creating and Modifying Variables
* Create new variable
gen log_income = log(income)
* Conditional assignment
gen high_earner = (income > 100000)
* Modify variable
replace age = . if age < 0
* Drop observations
drop if missing(income)
* Keep only certain variables
keep id income agelibrary(dplyr)
data <- data %>%
mutate(
# Create new variable
log_income = log(income),
# Conditional assignment
high_earner = income > 100000,
# Modify variable
age = if_else(age < 0, NA_real_, age)
) %>%
# Drop observations
filter(!is.na(income)) %>%
# Keep only certain variables
select(id, income, age)import numpy as np
# Create new variable
data["log_income"] = np.log(data["income"])
# Conditional assignment
data["high_earner"] = data["income"] > 100000
# Modify variable
data.loc[data["age"] < 0, "age"] = np.nan
# Drop observations
data = data.dropna(subset=["income"])
# Keep only certain variables
data = data[["id", "income", "age"]]Reshaping Data
Long Data is (Almost) Always Better
Most commands expect long format: multiple rows per unit, one column for values. If you're copy-pasting variable names with years (income_2018, income_2019...), you probably need to reshape to long.
Wide vs. Long Format
id income_2020 income_2021 1 50000 52000 2 60000 61000
One row per unit. Each time period is a separate column.
id year income 1 2020 50000 1 2021 52000 2 2020 60000 2 2021 61000
Multiple rows per unit. Two columns (id + year) together identify each row.
Key Insight: Multiple Identifier Columns
In long format, a single column no longer uniquely identifies each row. Instead, two or more columns together form the unique identifier:
- Panel data:
country+yeartogether identify each observation - Individual-time:
person_id+datetogether identify each observation - Repeated measures:
subject_id+treatment_roundtogether identify each observation
This is why Stata's reshape command requires both i() (the unit identifier) and j() (the new time/category variable).
Reshaping Wide to Long
* i() = unit identifier, j() = what becomes the new column
reshape long income_, i(id) j(year)
rename income_ incomelibrary(tidyr)
data_long <- data %>%
pivot_longer(
cols = starts_with("income_"),
names_to = "year",
names_prefix = "income_",
values_to = "income"
) %>%
mutate(year = as.numeric(year))# Melt wide to long
data_long = pd.melt(
data,
id_vars=["id"],
value_vars=["income_2020", "income_2021"],
var_name="year",
value_name="income"
)
# Clean up year column
data_long["year"] = data_long["year"].str.replace("income_", "").astype(int)Aggregating Data
Often you need to compute summary statistics within groups (by state, by year, etc.) or collapse data to a higher level of aggregation.
Group Calculations with egen
The egen command ("extensions to generate") creates new variables based on functions applied across observations:
* Mean across all observations
egen income_mean = mean(income)
* Mean within groups (by state)
bysort state: egen income_mean_state = mean(income)
* Other useful egen functions:
bysort state: egen income_max = max(income)
bysort state: egen income_sd = sd(income)
bysort state: egen income_count = count(income)
bysort state: egen income_p50 = pctile(income), p(50)library(dplyr)
# Add group-level statistics while keeping all rows
data <- data %>%
group_by(state) %>%
mutate(
income_mean_state = mean(income, na.rm = TRUE),
income_max = max(income, na.rm = TRUE),
income_sd = sd(income, na.rm = TRUE),
income_count = n(),
income_p50 = median(income, na.rm = TRUE)
) %>%
ungroup()# Add group-level statistics while keeping all rows
data["income_mean_state"] = data.groupby("state")["income"].transform("mean")
data["income_max"] = data.groupby("state")["income"].transform("max")
data["income_sd"] = data.groupby("state")["income"].transform("std")
data["income_count"] = data.groupby("state")["income"].transform("count")
data["income_p50"] = data.groupby("state")["income"].transform("median")gen sum vs. egen sum
In Stata, gen sum() creates a running total, while egen sum() creates the group total. Always read the help file—function names can be misleading!
Collapsing to Group Level
Use collapse to aggregate data to a higher level (e.g., individual → state, state-year → state):
* Collapse to state-level means
collapse (mean) income age, by(state)
* Multiple statistics
collapse (mean) income_mean=income ///
(sd) income_sd=income ///
(count) n=income, by(state)
* Different statistics for different variables
collapse (mean) income (max) tax_rate (sum) population, by(state year)# Collapse to state level
state_data <- data %>%
group_by(state) %>%
summarize(
income_mean = mean(income, na.rm = TRUE),
income_sd = sd(income, na.rm = TRUE),
n = n()
)
# Different statistics for different variables
state_year_data <- data %>%
group_by(state, year) %>%
summarize(
income = mean(income, na.rm = TRUE),
tax_rate = max(tax_rate, na.rm = TRUE),
population = sum(population, na.rm = TRUE),
.groups = "drop"
)# Collapse to state level
state_data = data.groupby("state").agg(
income_mean=("income", "mean"),
income_sd=("income", "std"),
n=("income", "count")
).reset_index()
# Different statistics for different variables
state_year_data = data.groupby(["state", "year"]).agg(
income=("income", "mean"),
tax_rate=("tax_rate", "max"),
population=("population", "sum")
).reset_index()egen vs. collapse
- egen — Adds a new column but keeps all rows. Use when you need the group statistic alongside individual data.
- collapse — Reduces the dataset to one row per group. Use when you want aggregated data only.
Merging Datasets
Merging combines two datasets that share a common identifier (like person ID or state-year).
Types of Joins
The most important thing to understand is what happens to rows that don't match:
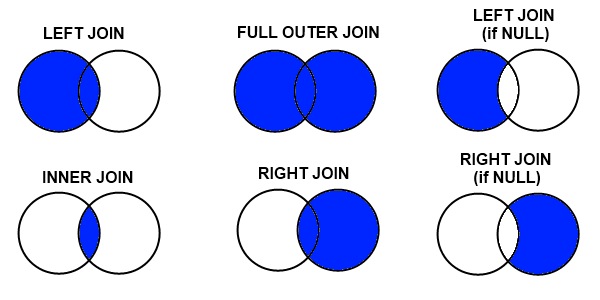
Credit: datacourses.com
- Inner join: Keep only rows that match in both datasets
- Left join: Keep all rows from the left (master) dataset
- Right join: Keep all rows from the right (using) dataset
- Full outer join: Keep all rows from both datasets
Merge Types by Relationship
Know Your Merge Type
- 1:1 — Each row matches exactly one row (person → demographics)
- m:1 — Many rows match one (students → school characteristics)
- 1:m — One row matches many (firm → firm-year panel)
- m:m — Almost always wrong! If you think you need this, reshape first.
When Merges Create Duplicates
Merges can increase your row count. This is sometimes expected and sometimes a bug.
Expected: m:1 or 1:m Merges
When merging individual data with aggregate data, rows should duplicate:
- 100 students + 5 schools → still 100 rows (each student gets their school's data)
- 50 states + 10 years of policy data → 500 state-year rows
Unexpected: Accidental Duplicates
If your row count increases unexpectedly, you probably have duplicate keys:
- Two "John Smith" entries in one dataset → each gets merged twice
- Forgot that some IDs appear multiple times
* BEFORE merging: Check for unexpected duplicates
duplicates report person_id // Should say "all unique" for 1:1
duplicates report state // OK to have duplicates for m:1
* Count rows before and after
count
local n_before = r(N)
merge m:1 state using "state_data.dta"
count
local n_after = r(N)
* If n_after > n_before in a m:1 merge, something is wrong!
di "Rows before: `n_before'"
di "Rows after: `n_after'"
if `n_after' > `n_before' {
di as error "WARNING: Row count increased - check for duplicate keys!"
}# BEFORE merging: Check for duplicates
individuals %>% count(person_id) %>% filter(n > 1) # Should be empty for 1:1
state_data %>% count(state) %>% filter(n > 1) # Should be empty
# Count rows before and after
n_before <- nrow(individuals)
merged <- individuals %>% left_join(state_data, by = "state")
n_after <- nrow(merged)
# Check if row count changed unexpectedly
if (n_after > n_before) {
warning("Row count increased! Check for duplicate keys in state_data")
# Find the problem
state_data %>% count(state) %>% filter(n > 1)
}# BEFORE merging: Check for duplicates
print(individuals.groupby("person_id").size().loc[lambda x: x > 1]) # Should be empty
print(state_data.groupby("state").size().loc[lambda x: x > 1]) # Should be empty
# Count rows before and after
n_before = len(individuals)
merged = individuals.merge(state_data, on="state", how="left")
n_after = len(merged)
# Check if row count changed unexpectedly
if n_after > n_before:
print("WARNING: Row count increased! Check for duplicate keys")
print(state_data.groupby("state").size().loc[lambda x: x > 1])The Row Count Rule
- 1:1 merge: Row count should stay exactly the same (or decrease if using inner join)
- m:1 merge: Row count should stay the same as master dataset
- 1:m merge: Row count will increase (one master row → many using rows)
- If row count increases when you didn't expect it, you have duplicate keys somewhere
Example: Merging Individual Data with State Data
Suppose you have individual-level data and want to add state characteristics:
person_id state income 1 MA 50000 2 MA 60000 3 CA 70000 4 NY 55000
state min_wage population MA 15.00 7000000 CA 15.50 39500000 TX 7.25 29500000
* Load master data
use "individuals.dta", clear
* Merge: many individuals per state → m:1
merge m:1 state using "state_data.dta"
* ALWAYS check the results
tab _merge
/*
_merge | Freq.
----------+------------
1 | 1 ← Only in master (NY - no state data)
2 | 1 ← Only in using (TX - no individuals)
3 | 3 ← Matched (MA, CA)
*/
* Investigate unmatched
list if _merge == 1 // NY had no state data
list if _merge == 2 // TX had no individuals
* Keep matched only (after understanding why others didn't match)
keep if _merge == 3
drop _mergelibrary(dplyr)
individuals <- read_dta("individuals.dta")
state_data <- read_dta("state_data.dta")
# Left join: keep all individuals, add state data where available
merged <- individuals %>%
left_join(state_data, by = "state")
# Check for unmatched
merged %>% filter(is.na(min_wage)) # NY had no state data
# Inner join: keep only matched
merged <- individuals %>%
inner_join(state_data, by = "state")individuals = pd.read_stata("individuals.dta")
state_data = pd.read_stata("state_data.dta")
# Left join: keep all individuals, add state data where available
merged = individuals.merge(state_data, on="state", how="left", indicator=True)
# Check merge results
print(merged["_merge"].value_counts())
# Check for unmatched
print(merged[merged["min_wage"].isna()]) # NY had no state data
# Inner join: keep only matched
merged = individuals.merge(state_data, on="state", how="inner")Pre-Merge Data Cleaning
Merge variables must be exactly identical in both datasets—same name, same values, same type. Most merge problems come from subtle differences that require string cleaning.
* Check if your variable is actually numeric or string
browse, nolabel // Shows actual values, not labels
* Common string cleaning operations:
* 1. Remove periods: "D.C." → "DC"
replace state = subinstr(state, ".", "", .)
* 2. Remove leading/trailing spaces: " MA " → "MA"
replace state = trim(state)
* 3. Extract substring: "Washington DC" → "DC"
replace state = substr(state, -2, 2) // Last 2 characters
* 4. Convert case: "ma" → "MA"
replace state = upper(state)
* 5. Rename to match other dataset
rename statefip state
* 6. Convert string to numeric (or vice versa)
destring state_code, replace
tostring state_fips, replacelibrary(stringr)
# Common string cleaning operations:
# 1. Remove periods
data$state <- str_replace_all(data$state, "\\.", "")
# 2. Remove leading/trailing spaces
data$state <- str_trim(data$state)
# 3. Extract substring
data$state <- str_sub(data$state, -2, -1) # Last 2 characters
# 4. Convert case
data$state <- str_to_upper(data$state)
# 5. Rename to match other dataset
data <- data %>% rename(state = statefip)
# 6. Convert types
data$state_code <- as.numeric(data$state_code)
data$state_fips <- as.character(data$state_fips)# Common string cleaning operations:
# 1. Remove periods
data["state"] = data["state"].str.replace(".", "", regex=False)
# 2. Remove leading/trailing spaces
data["state"] = data["state"].str.strip()
# 3. Extract substring
data["state"] = data["state"].str[-2:] # Last 2 characters
# 4. Convert case
data["state"] = data["state"].str.upper()
# 5. Rename to match other dataset
data = data.rename(columns={"statefip": "state"})
# 6. Convert types
data["state_code"] = pd.to_numeric(data["state_code"])
data["state_fips"] = data["state_fips"].astype(str)Labels Can Be Deceiving
In Stata, when you see "Washington" in the data browser, it might actually be stored as the number 53 with a label attached. Use browse, nolabel or tab var, nolabel to see the true values. Merges use the actual values, not the labels!
Common Merge Problems
Watch Out For
- Duplicates on key: If your key isn't unique when it should be, you'll get unexpected results. Always run
duplicates reportfirst. - Case sensitivity: "MA" ≠ "ma" in Stata. Use
upper()orlower()to standardize. - Trailing spaces: "MA " ≠ "MA". Use
trim()to remove. - Different variable types: String "1" ≠ numeric 1. Convert with
destringortostring. - Labels hiding values: What looks like "California" might be stored as 6. Check with
nolabel.
Appending (Stacking) Datasets
Use append to stack datasets vertically (same variables, different observations):
* Append multiple years of data
use "data_2020.dta", clear
append using "data_2021.dta"
append using "data_2022.dta"# Bind rows from multiple dataframes
data_all <- bind_rows(
read_dta("data_2020.dta"),
read_dta("data_2021.dta"),
read_dta("data_2022.dta")
)# Concatenate multiple dataframes
data_all = pd.concat([
pd.read_stata("data_2020.dta"),
pd.read_stata("data_2021.dta"),
pd.read_stata("data_2022.dta")
], ignore_index=True)Stata Traps
Stata has some behaviors that can silently corrupt your analysis.
Missing Values Are Infinity
In Stata, missing values (.) are treated as larger than any number. This means comparisons like if x > 100 will include missing values.
* This INCLUDES missing values (dangerous!)
gen high_income = (income > 100000)
* This is what you actually want
gen high_income = (income > 100000) if !missing(income)
* Or equivalently
gen high_income = (income > 100000 & income < .)
* The ordering is: all numbers < . < .a < .b < ... < .z
* So . > 999999999 evaluates to TRUE# R handles this more intuitively - NA propagates
high_income <- income > 100000
# Returns NA where income is NA, not TRUE
# But still be explicit about NA handling
high_income <- income > 100000 & !is.na(income)# Python/pandas handles this intuitively - NaN propagates
high_income = data["income"] > 100000
# Returns NaN where income is NaN, not True
# But still be explicit about NA handling
high_income = (data["income"] > 100000) & data["income"].notna()>, >=, or !=. Your sample sizes will be wrong, and your estimates will be biased.
egen group Silently Top-Codes
When you use egen group() to create a numeric ID from string or categorical variables, Stata uses an int by default. If you have more unique values than an int can hold (~32,000), it silently repeats the maximum value.
* DANGEROUS: If you have >32,767 unique beneficiaries,
* later IDs will all be assigned the same number!
egen bene_id = group(beneficiary_name)
* SAFE: Explicitly use a long integer
egen long bene_id = group(beneficiary_name)
* Or for very large datasets (>2 billion unique values)
egen double bene_id = group(beneficiary_name)
* Always verify you got the right number of groups
distinct beneficiary_name
assert r(ndistinct) == r(N) // This will fail if top-coded# R doesn't have this problem - factors work fine
data$bene_id <- as.numeric(factor(data$beneficiary_name))
# Or use dplyr
data <- data %>%
mutate(bene_id = as.numeric(factor(beneficiary_name)))# Python doesn't have this problem - uses 64-bit integers by default
data["bene_id"] = pd.factorize(data["beneficiary_name"])[0] + 1
# Verify
assert data["bene_id"].nunique() == data["beneficiary_name"].nunique()Numeric Precision
Stata's float type only has ~7 digits of precision. Large IDs stored as floats will be silently corrupted.
* float has ~7 digits of precision
* This can cause merge failures on IDs!
gen float id = 1234567890
* id is now 1234567936 (corrupted!)
* Always use long or double for IDs
gen long id = 1234567890 // integers up to ~2 billion
gen double id = 1234567890 // integers up to ~9 quadrillion
* Check if a variable might have precision issues
summarize id
* If max is > 16777216 and storage type is float, you have a problem
* Fix: compress optimizes storage types automatically
compress# R uses double precision by default, so this is less of an issue
# But be careful with very large integers
id <- 1234567890 # Fine in R
# For extremely large integers, use bit64 package
library(bit64)
big_id <- as.integer64("9223372036854775807")# Python uses 64-bit integers by default, so this is less of an issue
id = 1234567890 # Fine in Python
# pandas also handles large integers well
data["id"] = pd.to_numeric(data["id"], downcast=None) # Keep full precision
# Check dtypes
print(data.dtypes)Defensive Coding Habits
- Always use
if !missing(x)with inequality comparisons - Always use
egen longoregen doublefor group IDs - Run
compressafter loading data to optimize storage types - Use
assertliberally to catch problems early